 Author
Author  Correspondence author
Correspondence author
Maize Genomics and Genetics, 2024, Vol. 15, No. 3 doi: 10.5376/mgg.2024.15.0015
Received: 10 May, 2024 Accepted: 12 Jun., 2024 Published: 30 Jun., 2024
Li Z., 2024, Transposons in zea genomics: their impact on genetic architecture, Maize Genomics and Genetics, 15(3): 147-159 (doi: 10.5376/mgg.2024.15.0015)
Transposons, also known as mobile genetic elements, play a significant role in shaping the genetic architecture of Zea species. This review delves into the diverse types of transposons present in Zea genomes, including DNA transposons and retrotransposons, and their mechanisms of action. We explore how these elements contribute to genetic variation, genome evolution, and the regulation of gene expression. The review also highlights the evolutionary forces that influence the maintenance and diversification of transposons within Zea genomes. Furthermore, we discuss the implications of transposon activity for plant breeding and genetic research, emphasizing their potential as tools for functional genomics and the development of new cultivars. By synthesizing current knowledge, this review provides a comprehensive understanding of the impact of transposons on the genetic architecture of Zea species.
1 Introduction
The genus Zea comprises several species, with maize (Zea mays ssp. mays) and its wild ancestor teosinte (Zea mays ssp. parviglumis) being the most notable. Maize, a staple crop with significant economic and nutritional value, was domesticated from teosinte approximately 9 000 years ago in southern Mexico (Xu et al., 2019; Li et al., 2021). Teosinte, which still grows wild in regions of Mexico, exhibits considerable genetic diversity and resilience to various biotic and abiotic stresses (Adhikari et al., 2021). The evolutionary transition from teosinte to maize involved substantial morphological and genetic changes, including alterations in plant architecture, seed size, and metabolic pathways (Dorweiler and Doebley, 1997; Dermastia et al., 2009; Xu et al., 2019).
Transposons, or transposable elements, are DNA sequences that can change their position within the genome, thereby influencing genetic diversity and evolution. In plant genomics, transposons play a crucial role in shaping the genetic architecture by inducing mutations, altering gene expression, and contributing to genome size variation (Tian et al., 2019; Li et al., 2021). In maize and teosinte, transposons have been implicated in significant genomic rearrangements and the evolution of key traits that distinguish domesticated maize from its wild ancestor (Dorweiler and Doebley, 1997; Li et al., 2021). Understanding the impact of transposons is essential for unraveling the complexities of plant evolution, adaptation, and breeding.
This study provides a comprehensive analysis of the role of transposons in the genomics of Zea species, focusing on their impact on genetic architecture. By synthesizing current research findings, elucidates how transposons have contributed to the genetic divergence between maize and teosinte and their implications for plant breeding and genetic improvement. The significance of this study lies in its potential to enhance our understanding of plant genome evolution and to inform strategies for utilizing wild germplasm in crop improvement programs. This study hopes to highlight the importance of transposons as drivers of genetic innovation and their potential applications in modern agriculture.
2 Historical Perspective of Transposon Research in Zea
2.1 Discovery of transposons by Barbara McClintock
The discovery of transposons, or "jumping genes," by Barbara McClintock in the late 1940s revolutionized our understanding of genetic elements and their behavior. McClintock's meticulous studies on chromosome breakage in maize led her to identify a chromosome-breaking locus that could change its position within a chromosome. This groundbreaking work challenged the prevailing notion of genes as stable entities arranged linearly on chromosomes, akin to beads on a string (Ravindran, 2012). McClintock's discovery of the Ac (Activator) and Ds (Dissociation) elements, which could move within the genome and alter gene expression depending on their insertion sites, laid the foundation for the field of transposon research (Ravindran, 2012).
Despite initial skepticism, McClintock's findings were gradually accepted by maize geneticists, and her work gained wider recognition over time. The significance of her discovery was eventually acknowledged with numerous prestigious awards, including the 1983 Nobel Prize in Physiology or Medicine. Her pioneering research not only unveiled the dynamic nature of the genome but also highlighted the potential of transposons as tools for genetic studies and manipulation (Ravindran, 2012).
2.2 Early studies on maize transposons
Following McClintock's discovery, the 1950s and 1960s saw a surge in research activity focused on transposable elements in maize. Researchers began to explore the genetic and molecular mechanisms underlying transposition and its effects on gene expression. One of the early significant contributions was the identification of the cut-and-paste mechanism of transposition by the R.A. Brink lab, which demonstrated how transposons could move to nearby sites on the chromosome (Peterson, 2005). This period also saw the description of various transposable element systems, such as the En/Spm (Enhancer/Suppressor-mutator) system, which further expanded our understanding of the diversity and functionality of transposons in maize (Peterson, 2005).
As molecular techniques advanced, researchers in the 1980s began to isolate and characterize the structure and size of different transposable elements. This molecular exploration revealed that over 75% of the maize genome consists of mobile elements, underscoring the profound impact of transposons on the genetic architecture of maize (Peterson, 2005). These early studies laid the groundwork for subsequent research that leveraged transposons as tools for gene discovery and functional genomics.
2.3 Evolution of research techniques and methodologies
The evolution of research techniques and methodologies has significantly advanced our understanding of transposons and their applications in maize genomics. The development of transposon-tagging strategies has been instrumental in gene discovery and functional studies. For instance, the use of Mutator (Mu) transposons in maize has enabled the identification of gene locations through genome resequencing and the isolation of specific genes, such as the lazy plant1 (la1) gene, which is involved in gravitropism. This method combines restriction enzyme digestion with high-throughput sequencing to map transposon insertion sites, facilitating the study of gene function and regulation (Howard et al., 2014).
In addition to transposon-tagging, advancements in epigenetic research have shed light on the regulation of transposon activity. Studies have shown that transposon silencing is associated with DNA methylation, histone modifications, and RNA interference (RNAi) pathways. These epigenetic mechanisms play crucial roles in maintaining genome stability and preventing the mutagenic effects of transposon insertions (Lippman et al., 2003). The integration of genetic and epigenetic approaches has provided a comprehensive understanding of transposon behavior and its implications for genome evolution and function.
The historical perspective of transposon research in Zea highlights the transformative impact of McClintock's discovery and the subsequent advancements in research techniques. From the initial identification of mobile elements to the development of sophisticated methodologies for gene discovery and epigenetic regulation, the study of transposons in maize continues to be a dynamic and evolving field with far-reaching implications for genetics and genomics.
3 Types of Transposons in Zea
3.1 Classification of transposons
Transposons, also known as "jumping genes", are DNA sequences capable of moving from one location to another within a genome. They are broadly classified into two main categories: DNA transposons and retrotransposons. DNA transposons, also known as Class II transposons, move directly as DNA through a "cut-and-paste" mechanism. This involves the excision of the transposon from one genomic location and its integration into another, facilitated by the enzyme transposase (Johnson and Reznikoff, 1983; Feschotte and Pritham, 2007; Hickman and Dyda, 2016). Retrotransposons, or Class I transposons, move via an RNA intermediate. This process involves transcription of the transposon into RNA, reverse transcription into DNA, and integration of the new DNA copy into the genome. Retrotransposons are further divided into long terminal repeat (LTR) and non-LTR retrotransposons (Hancks and Kazazian, 2012).
In Zea mays (maize), both DNA transposons and retrotransposons are prevalent and play significant roles in genome evolution and diversity. The maize genome is particularly rich in transposable elements, which constitute a substantial portion of its genetic material. The classification of these elements is crucial for understanding their impact on the genetic architecture of maize, as different types of transposons have distinct mechanisms of transposition and genomic effects (Feschotte and Pritham, 2007; Huang et al., 2012; Burns, 2020).
3.2 Key transposon families
Among the various transposon families in Zea, the most notable include the Activator (Ac)/Dissociation (Ds) system, the Mutator (Mu) family, and the Helitron family. The Ac/Ds system is a well-studied DNA transposon family in maize. The Ac element encodes a transposase that facilitates its own movement as well as the movement of non-autonomous Ds elements, which lack functional transposase genes (Johnson and Reznikoff, 1983; Feschotte and Pritham, 2007). This system has been instrumental in studying gene function and regulation in maize.
The Mutator family is another significant group of DNA transposons in maize. These elements are highly mutagenic and have been extensively used in genetic studies to induce mutations and identify gene functions. The Mu elements are characterized by their high transposition activity and ability to generate a wide range of genetic variations (Feschotte and Pritham, 2007; Huang et al., 2012).
Helitrons represent a unique family of rolling-circle transposons that have been identified in maize. Unlike other DNA transposons, Helitrons replicate through a rolling-circle mechanism, which involves the formation of a single-stranded DNA intermediate. This family of transposons is known for capturing and shuffling gene fragments, thereby contributing to genome plasticity and evolution (Feschotte and Pritham, 2007; Hickman and Dyda, 2012).
3.3 Mechanisms of transposition
The mechanisms of transposition vary among different types of transposons, but they generally involve a series of well-coordinated steps. For DNA transposons, the process typically begins with the recognition of specific DNA sequences at the ends of the transposon by the transposase enzyme. The transposase then catalyzes the excision of the transposon from its original location and its integration into a new genomic site. This "cut-and-paste" mechanism is characteristic of many DNA transposons, including the Ac/Ds and Mu elements in maize (Johnson and Reznikoff, 1983; Feschotte and Pritham, 2007; Hickman and Dyda, 2016).
Retrotransposons, on the other hand, utilize a "copy-and-paste" mechanism. The transposition process starts with the transcription of the retrotransposon into RNA, followed by reverse transcription into DNA by the enzyme reverse transcriptase. The newly synthesized DNA copy is then integrated into a new genomic location. This mechanism is typical of LTR and non-LTR retrotransposons, which are abundant in the maize genome (Fedoroff, 2012; Hancks and Kazazian, 2012).
Helitrons employ a distinct rolling-circle replication mechanism. The process involves the formation of a single-stranded DNA intermediate, which is then used as a template for the synthesis of a new double-stranded DNA copy. This new copy is subsequently integrated into the genome. The unique mechanism of Helitrons allows them to capture and mobilize gene fragments, contributing to genetic diversity and innovation in maize (Feschotte and Pritham, 2007; Hancks and Kazazian, 2012). The diverse mechanisms of transposition employed by different transposon families in Zea mays highlight the complexity and dynamism of the maize genome. Understanding these mechanisms is essential for elucidating the roles of transposons in genome evolution, gene regulation, and genetic diversity in maize.
4 Transposon Distribution and Abundance in Zea Genomes
4.1 Genome-wide distribution patterns
Transposable elements (TEs) are a significant component of the maize genome, constituting over 85% of its DNA sequence (Figure 1) (Stitzer et al., 2019). These elements are not uniformly distributed across the genome; instead, they exhibit distinct patterns of localization. For instance, certain TIR (Terminal Inverted Repeat) elements are less prevalent in centromeric and pericentromeric regions, while others do not show such biases (Su et al., 2019). This uneven distribution suggests that TEs may have specific insertion preferences or that certain genomic regions are more permissive to TE insertions.
.png) Figure 1 Chromosomal distribution of superfamilies and example families (Adopted from Stitzer et al., 2019) Image caption: Counts of number of insertions in 1 Mb bins across chromosome 1 for (A) TE superfamilies and (B-E) the 5 families with highest copy number in each of four superfamilies, DHH (B), DTT (C), RLC (D), and RLG (E) (Adopted from Stitzer et al., 2019) |
The dynamic nature of TE distribution is further highlighted by the observation that different TE families exhibit tissue-specific expression patterns. For example, a significant number of TE families are specifically active in pollen and endosperm, which are critical for reproductive processes (Anderson et al., 2019). This tissue-specific activity indicates that TEs may play roles in developmental regulation and genome evolution, contributing to the genetic diversity observed in maize.
4.2 Comparative analysis between maize and teosinte
Comparative genomic studies between maize (Zea mays L.) and its wild progenitor, teosinte (Zea mays ssp. parviglumis), reveal significant differences in TE content and distribution. Teosinte, which serves as an important genetic resource for maize improvement, has a distinct TE landscape compared to domesticated maize. For instance, genes in teosinte exhibit more transcript isoforms and are enriched in RNA modification pathways, suggesting a more complex regulatory network influenced by TEs (Li et al., 2021a).
Moreover, the intergenic regions in teosinte are extensively altered by TEs, indicating that these elements have played a crucial role in the genomic divergence between maize and teosinte (Li et al., 2021a). This divergence is not only a result of TE insertions but also due to the differential retention and amplification of specific TE families. Such comparative analyses underscore the evolutionary impact of TEs on the genetic architecture of Zea species.
4.3 Methods used for identifying and mapping transposons
Several advanced methodologies have been developed to identify and map TEs in the maize genome. One such method involves the use of single-molecule long-read sequencing, which allows for the accurate annotation of TEs without the need for a reference genome (Li et al., 2021a). This approach has been particularly useful in characterizing the TE landscape in teosinte, providing insights into the genomic changes that accompanied maize domestication.
Another effective technique is the use of a capture-based assay, which targets specific transposon polymorphisms across various maize genotypes. This method has demonstrated high reliability, with a consistency rate of 98.6% when compared to PCR-based assays (Li et al., 2021b). By integrating transposon polymorphism data with gene expression profiles, researchers can identify TEs that influence gene expression, thereby elucidating their functional roles in the genome.
Additionally, bioinformatics tools such as TIR-Learner have been developed to enhance the detection and annotation of TIR elements. This ensemble method combines homology-based and de novo machine-learning approaches, significantly improving the accuracy and efficiency of TIR element annotation (Su et al., 2019). Such tools are essential for understanding the full extent of TE diversity and their impact on genome structure and function.
The distribution and abundance of TEs in Zea genomes are shaped by a combination of evolutionary processes and methodological advancements. Comparative analyses between maize and teosinte highlight the role of TEs in genome evolution, while innovative techniques for TE identification and mapping continue to refine our understanding of their contributions to genetic diversity.
5 Impact of Transposons on Genetic Architecture
5.1 Gene expression regulation
Transposons, also known as transposable elements (TEs), play a significant role in the regulation of gene expression. These mobile genetic elements can act as sources of transcriptional modulatory elements, such as gene promoters and enhancers, splicing and termination sites, and regulatory non-coding RNAs. This ability allows transposons to influence the expression of nearby genes, either enhancing or repressing their activity (Elbarbary et al., 2016; Branco and Chuong, 2020). For instance, the insertion of a transposon near a gene can introduce new regulatory sequences that alter the gene's expression pattern, potentially leading to novel phenotypic traits.
Moreover, transposons have driven the evolution of host defense mechanisms that have been repurposed for gene regulation. These mechanisms include the silencing of transposons through DNA methylation and histone modification, which can also affect the expression of nearby genes. The interplay between transposons and gene regulation is complex and context-dependent, often requiring specialized analytical tools to dissect their functional roles (Figure 2) (Branco and Chuong, 2020; Bhat et al., 2022).
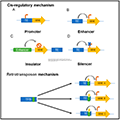 Figure 2 TE-regulated mechanisms of action in the host cells (Adopted from Bhat et al., 2022) Image caption: Cis-regulatory mechanisms involving (A) promoter and (B) enhancer, integrate the activity of specific transcription factor; (C) insulator, act either through enhancer-blocking activity or chromatin barrier activity; (D) silencer, silence the expression of genes; Retrotransposon mechanism can increase the potential of transcription binding factor. (orange arrowhead indicates increased activity, blue cross indicates silencing of activity, circle with single cross indicates insulation of gene activity, grey arrow indicates direction of action) (Adopted from Bhat et al., 2022) |
5.2 Genome size and structure
Transposons contribute significantly to the size and structure of genomes. In maize (Zea mays), for example, transposons make up a substantial portion of the genome, with estimates suggesting that they constitute up to 90% of the total genomic content (Xue and Goldenfeld, 2016). The insertion and subsequent amplification of transposons can lead to large-scale genomic alterations, including inversions, deletions, and duplications, which can have profound effects on genome architecture (Bhat et al., 2022).
The presence of transposons can also lead to the formation of new genomic regions with distinct structural features. For instance, the insertion of transposons can create new sites for chromatin remodeling, thereby influencing the overall organization of the genome. This dynamic restructuring of the genome by transposons is a key driver of genomic diversity and evolution (Elbarbary et al., 2016; Bhat et al., 2022).
In addition to generating genetic diversity through insertional mutagenesis, transposons can also facilitate the horizontal transfer of genetic material between species. This horizontal gene transfer can introduce new genes and regulatory elements into a genome, further enhancing genetic diversity and evolutionary potential. The dual role of transposons as both creators of genetic variation and facilitators of gene transfer underscores their importance in the evolutionary process (Percharde et al., 2020; Bhat et al., 2022).
5.4 Specific examples of transposon-induced mutations
One notable example of a transposon-induced mutation is the insertion of the transposable element Ac (Activator) in the maize genome, which can cause the breakage of chromosomes and lead to variegated phenotypes in maize kernels. This phenomenon was first described by Barbara McClintock and has since been recognized as a classic example of transposon activity influencing genetic traits (Perween et al., 2020).
Another example is the role of the Long INterspersed Element-1 (LINE-1 or L1) in human disease. LINE-1 is a type of non-LTR retrotransposon that is currently active in humans and has been implicated in various genetic disorders. For instance, the insertion of LINE-1 elements can disrupt gene function and lead to diseases such as hemophilia and Duchenne muscular dystrophy. The ongoing activity of LINE-1 in the human genome highlights the potential for transposons to cause both beneficial and deleterious mutations (Hancks and Kazazian, 2012).
Transposons have a profound impact on the genetic architecture of organisms. They regulate gene expression, shape genome size and structure, generate genetic diversity, and can induce specific mutations that influence phenotypic traits. Understanding the multifaceted roles of transposons is crucial for unraveling the complexities of genome evolution and function.
6 Transposons and Epigenetic Regulation
6.1 Role of transposons in epigenetic modifications
Transposons, also known as transposable elements (TEs), are mobile genetic elements that can move within a genome and have a significant impact on its structure and function. One of the primary ways transposons influence the genome is through epigenetic modifications. These modifications include DNA methylation and histone modifications, which can alter gene expression without changing the underlying DNA sequence. Transposons can serve as sites for these epigenetic marks, thereby influencing the regulation of nearby genes. For instance, transposons have been shown to be involved in the formation of new regulatory regions, such as promoters and enhancers, which can drive the evolution of gene regulatory networks (Friedli and Trono, 2015; Igolkina et al., 2019).
The role of transposons in epigenetic regulation is not limited to the addition of epigenetic marks. They also play a crucial role in the dynamic regulation of these marks during development and in response to environmental changes. For example, during embryonic development, transposons are tightly regulated by epigenetic mechanisms to ensure proper gene expression patterns. This regulation is achieved through a combination of DNA methylation and histone modifications, which work together to silence transposons and prevent their potentially deleterious effects on the genome (Huda and Jordan, 2009; Friedli and Trono, 2015).
6.2 Interaction with DNA methylation and histone modification
Transposons interact with various epigenetic mechanisms, including DNA methylation and histone modifications, to regulate their activity and impact on the genome. DNA methylation, particularly at CpG sites, is a well-known mechanism for silencing transposons. This methylation can prevent the transcription of transposon sequences, thereby reducing their ability to move and insert into new genomic locations. Studies have shown that mutations in DNA methyltransferases, such as MET1, can lead to the reactivation of transposons, highlighting the importance of DNA methylation in transposon silencing (Lippman et al., 2003; Mustafin and Khusnutdinova, 2018).
Histone modifications also play a critical role in the regulation of transposons. Specific histone marks, such as H3K9me3 and H3K27me3, are associated with the formation of heterochromatin, a tightly packed form of DNA that is transcriptionally inactive. These histone marks can be found at transposon sequences, contributing to their silencing. For example, in Xenopus tropicalis embryos, different types of transposons are marked by distinct combinations of heterochromatic histone modifications, which vary depending on the developmental stage and the type of transposon (Kruijsbergen et al., 2017). This suggests that histone modifications are a key component of the epigenetic regulation of transposons.
Histone modifications also play a critical role in the regulation of transposons. Specific histone marks, such as H3K9me3 and H3K27me3, are associated with the formation of heterochromatin, a tightly packed form of DNA that is transcriptionally inactive. These histone marks can be found at transposon sequences, contributing to their silencing. For example, in Xenopus tropicalis embryos, different types of transposons are marked by distinct combinations of heterochromatic histone modifications, which vary depending on the developmental stage and the type of transposon (Kruijsbergen et al., 2017). This suggests that histone modifications are a key component of the epigenetic regulation of transposons.
Another case study in maize involves the role of transposons in the regulation of gene expression during development. Transposons can influence the expression of nearby genes by serving as sites for epigenetic marks. For example, the insertion of a transposon near a gene can lead to the recruitment of DNA methylation and histone modifications, which can either activate or repress the gene depending on the specific marks involved. This dynamic regulation allows for the fine-tuning of gene expression in response to developmental cues and environmental changes (Weil and Martienssen, 2008; Mustafin and Khusnutdinova, 2018).
Transposons play a crucial role in the epigenetic regulation of the genome in Zea mays. Through interactions with DNA methylation and histone modifications, transposons can influence gene expression and contribute to the evolution of gene regulatory networks. These interactions highlight the complex and dynamic nature of epigenetic regulation and the important role of transposons in shaping the genetic architecture of maize.
7 Transposons in Zea Breeding and Genetic Enhancement
7.1 Utilization of transposons as molecular markers
Transposons, or transposable elements, are mobile genetic sequences that can move within a genome, making them valuable tools in genetic research and breeding. In Zea mays (maize), transposons have been effectively utilized as molecular markers due to their ability to create unique insertion patterns within the genome. These patterns, referred to as transposon signatures, can be used to differentiate between closely related species and subspecies, providing a robust method for phylogenetic and population genetic studies (Purugganan and Wessler, 1995). The PCR-based method developed to utilize these transposon signatures allows for the examination of relationships within the genus Zea, making it a powerful tool for genetic mapping and marker-assisted selection in breeding programs.
Moreover, the use of transposons as molecular markers is not limited to differentiation between species. They also play a crucial role in identifying genetic variations within a species. For instance, the magellan retrotransposon has been used to generate transposon signatures that help in understanding the genetic diversity and evolutionary relationships within maize populations (Purugganan and Wessler, 1995). This capability is particularly useful in breeding programs aimed at enhancing specific traits, as it allows breeders to track the inheritance of desirable genes and select for individuals with optimal genetic combinations.
7.2 Role in trait improvement and hybrid vigor
Transposons contribute significantly to trait improvement and hybrid vigor in maize by facilitating genetic diversity and enabling the introduction of new genetic variations. These mobile elements can insert themselves into various genomic locations, potentially disrupting or enhancing gene function. This process can lead to the creation of novel phenotypes, which can be harnessed for trait improvement. For example, transposon insertional mutagenesis has been used to generate gain-of-function phenotypes in maize, which are valuable for studying gene function and improving crop traits (Qu et al., 2007).
The activation of transposons can also play a role in hybrid vigor, or heterosis, which is the phenomenon where hybrid offspring exhibit superior qualities compared to their parents. Hybridization between genetically diverse maize lines can trigger transposon mobilization, leading to genomic reorganization and the creation of new genetic combinations. This genomic shock can result in the activation of previously silent transposons, contributing to the genetic and phenotypic diversity observed in hybrid maize populations (Fukai et al., 2022). The transgenerational activation of transposons, as seen in other plant species, suggests that these elements can have long-term effects on the genetic architecture of hybrids, further enhancing their vigor and adaptability (Fukai et al., 2022).
7.3 Case studies of successful breeding programs leveraging transposons
Several breeding programs have successfully leveraged transposons to enhance maize genetics and improve crop traits. One notable example is the use of the Ac-Ds transposon system in maize. This system involves the use of a Dissociation (Ds) element and an Activator (Ac) transposase gene to induce transposon mobilization. Researchers have developed an activation-tagging vector system using Ac-Ds, which has been tested in rice and shown to be effective in generating stable transposon insertions (Qu et al., 2007). The success of this system in rice suggests its potential applicability in maize, where it can be used to create novel phenotypes and improve crop traits through targeted gene activation.
Another successful case study involves the use of transposons in functional genomics to identify and characterize genes associated with important agronomic traits. For instance, the Sleeping Beauty transposon system has been used in vertebrate models to induce mutations and study gene function (Kawakami et al., 2017). Similar approaches can be applied in maize to create loss-of-function and gain-of-function mutations, providing insights into the genetic basis of traits such as disease resistance, yield, and stress tolerance. By integrating transposon-based mutagenesis with traditional breeding techniques, researchers can accelerate the development of maize varieties with enhanced performance and resilience.
In conclusion, transposons play a pivotal role in maize breeding and genetic enhancement by serving as molecular markers, contributing to trait improvement and hybrid vigor, and enabling the development of innovative breeding strategies. The successful application of transposon-based technologies in other plant species and model organisms underscores their potential to revolutionize maize breeding and drive the creation of superior crop varieties.
8 Future Prospects and Challenges
8.1 Advances in genomic technologies and their impact on transposon research
Recent advancements in genomic technologies have significantly enhanced our understanding of transposons and their roles in shaping the genetic architecture of Zea mays. High-throughput sequencing technologies, such as next-generation sequencing (NGS), have enabled the comprehensive mapping of transposon insertions across the genome, providing insights into their distribution, activity, and evolutionary impact (Feschotte and Pritham, 2007; Huang et al., 2012). These technologies have also facilitated the identification of active transposons, which continue to create new insertions and contribute to genetic diversity (Huang et al., 2012). Additionally, the development of CRISPR-Cas9 and other genome-editing tools has opened new avenues for studying the functional roles of transposons by allowing precise manipulation of their sequences and insertion sites (Sandoval-Villegas et al., 2021).
Moreover, bioinformatics tools and computational models have become indispensable in transposon research. These tools enable the analysis of large genomic datasets, helping researchers to predict transposon behavior, identify potential regulatory elements, and understand the mechanisms underlying transposon-mediated gene regulation (Bhat et al., 2022). As these technologies continue to evolve, they will likely uncover new aspects of transposon biology and their contributions to genome evolution and stability.
8.2 Potential applications in biotechnology and crop improvement
Transposons hold great promise for biotechnological applications and crop improvement. One of the most significant applications is in the field of functional genomics, where transposons are used as tools for insertional mutagenesis to identify and characterize gene functions (Ramachandran and Sundaresan, 2001). This approach has been successfully employed in various plant species, including maize, to generate mutant libraries and study gene function on a genome-wide scale (Qu et al., 2007). Additionally, transposons can be used as activation tags to create gain-of-function mutations, which can help identify genes involved in important agronomic traits (Qu et al., 2007).
In crop improvement, transposons can be harnessed to introduce beneficial traits, such as disease resistance, stress tolerance, and improved yield. For example, the Ac-Ds transposon system has been used to enhance gene expression in rice, demonstrating its potential for improving crop performance (Qu et al., 2007). Furthermore, transposons can be employed in gene editing strategies to create targeted modifications in the genome, offering a powerful tool for precision breeding and the development of genetically modified crops with desirable traits (Sandoval-Villegas et al., 2021).
In crop improvement, transposons can be harnessed to introduce beneficial traits, such as disease resistance, stress tolerance, and improved yield. For example, the Ac-Ds transposon system has been used to enhance gene expression in rice, demonstrating its potential for improving crop performance (Qu et al., 2007). Furthermore, transposons can be employed in gene editing strategies to create targeted modifications in the genome, offering a powerful tool for precision breeding and the development of genetically modified crops with desirable traits (Sandoval-Villegas et al., 2021).
Ecologically, the release of genetically modified organisms (GMOs) containing transposons into the environment could have significant impacts on biodiversity and ecosystem stability. Transposons have the potential to spread horizontally between species, which could lead to the unintended transfer of genetic material and the creation of new transposon insertions in wild populations (Bhat et al., 2022). To mitigate these risks, it is essential to develop containment strategies and monitor the ecological effects of transposon-based GMOs.
8.4 Challenges in transposon research and possible solutions
Despite the advancements in transposon research, several challenges remain. One of the main challenges is the difficulty in predicting transposon behavior and insertion sites, which can complicate the interpretation of experimental results and the development of transposon-based applications (Feschotte and Pritham, 2007). To address this issue, researchers are developing more sophisticated bioinformatics tools and computational models to improve the accuracy of transposon predictions and enhance our understanding of their mechanisms (Bhat et al., 2022).
Another challenge is the potential for transposon-induced genomic instability, which can lead to deleterious mutations and affect the overall fitness of the organism (Bhat et al., 2022). To overcome this, researchers are exploring strategies to control transposon activity, such as the use of inducible promoters and targeted genome editing techniques to minimize unintended insertions and ensure precise modifications (Sandoval-Villegas et al., 2021). Additionally, ongoing efforts to characterize the regulatory networks and epigenetic mechanisms that govern transposon activity will provide valuable insights into how to harness their potential while mitigating their risks.
While transposon research in Zea genomics presents exciting opportunities for scientific discovery and practical applications, it also poses significant challenges that must be carefully addressed. Continued advancements in genomic technologies, coupled with a thorough understanding of the ethical and ecological implications, will be essential for realizing the full potential of transposons in biotechnology and crop improvement.
9 Concluding Remarks
Transposons, or transposable elements (TEs), are significant contributors to the genetic architecture of Zea species. They are mobile genetic units that can move within the genome, causing mutations, chromosomal rearrangements, and influencing gene expression. Studies have shown that TEs are selectively retained near genes involved in environmental adaptation, such as xenobiotic-metabolizing cytochrome P450 genes in Helicoverpa zea. TEs play a crucial role in genome evolution by creating genetic diversity and driving structural changes in the genome. In maize, TEs have been shown to induce complex chromosomal rearrangements, which can lead to significant genetic variation and influence gene expression. Additionally, TEs contribute to the evolution of the genome by both active and passive mechanisms, impacting genome stability and gene regulation.
Continued research on transposons in Zea genomics is essential for several reasons. Understanding the mechanisms by which TEs influence genome architecture can provide insights into the evolutionary processes that shape genetic diversity in Zea species. Studying the impact of TEs on gene regulation can reveal how these elements contribute to the adaptation of Zea species to their environments. Research on TEs can uncover the potential for using these elements in genetic engineering and crop improvement, as they can be harnessed to introduce beneficial traits or enhance genetic diversity. Investigating the role of TEs in genome stability and their potential implications for health and disease can lead to the development of new strategies for managing genetic disorders and improving crop resilience.
The future of transposon research in plant genomics holds great promise. Advances in genomic technologies and analytical tools will enable more precise characterization of TEs and their effects on the genome. As researcher’s understanding of the complex interactions between TEs and the host genome deepens, can expect to uncover new mechanisms by which these elements drive genetic innovation and adaptation. Furthermore, the potential applications of TEs in genetic engineering and crop improvement are vast, offering opportunities to enhance crop yields, improve resistance to pests and diseases, and adapt to changing environmental conditions. Overall, continued research on transposons will not only advance researcher’s knowledge of genome evolution but also provide practical solutions for addressing global agricultural challenges.
Acknowledgments
The author extends sincere thanks to two anonymous peer reviewers for their feedback on the manuscript.
Conflict of Interest Disclosure
The author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Adhikari S., Joshi A., Kumar A., and Singh N., 2021, Diversification of maize (Zea mays L.) through teosinte (Zea mays subsp. parviglumis Iltis & Doebley) allelic, Genetic Resources and Crop Evolution, 68: 2983-2995.
https://doi.org/10.1007/s10722-021-01170-z
Anderson S., Stitzer M., Zhou P., Ross-Ibarra J., Hirsch C., and Springer N., 2019, Dynamic patterns of transcript abundance of transposable element families in maize, G3: Genes|Genomes|Genetics, 9: 3673-3682.
https://doi.org/10.1534/g3.119.400431
PMid:31506319 PMCid:PMC6829137
Bhat A., Ghatage T., Bhan S., Lahane G., Dhar A., Kumar R., Pandita R., Bhat K., Ramos K., and Pandita T., 2022, Role of transposable elements in genome stability: implications for health and disease, International Journal of Molecular Sciences, 23(14): 7802.
https://doi.org/10.3390/ijms23147802
PMid:35887150 PMCid:PMC9319628
Branco M., and Chuong E., 2020, Crossroads between transposons and gene regulation, Philosophical Transactions of the Royal Society B, 375(1795): 20190330.
https://doi.org/10.1098/rstb.2019.0330
PMid:32075561 PMCid:PMC7061990
Burns K., 2020, Our conflict with transposable elements and its implications for human disease, Annual Review of Pathology, 15: 51-70.
https://doi.org/10.1146/annurev-pathmechdis-012419-032633
PMid:31977294
Dermastia M., Kladnik A., Koce J., and Chourey P., 2009, A cellular study of teosinte Zea mays subsp. parviglumis (Poaceae) caryopsis development showing several processes conserved in maize, American Journal of Botany, 96(10): 1798-1807.
https://doi.org/10.3732/ajb.0900059
PMid:21622300
Dorweiler J., and Doebley J., 1997, Developmental analysis of teosinte glume architecture1: a key locus in the evolution of maize (Poaceae), American Journal of Botany, 84(10): 1313.
https://doi.org/10.2307/2446130
PMid:21708541
Elbarbary R., Lucas B., and Maquat L., 2016, Retrotransposons as regulators of gene expression, Science, 351(6274): aac7247.
https://doi.org/10.1126/science.aac7247
PMid:26912865 PMCid:PMC4788378
Fedoroff N., 2012, Transposable elements, epigenetics, and genome evolution, Science, 338: 758-767.
https://doi.org/10.1126/SCIENCE.338.6108.758
PMid:23145453
Feschotte C., and Pritham E., 2007, DNA transposons and the evolution of eukaryotic genomes, Annual Review of Genetics, 41: 331-368.
https://doi.org/10.1146/ANNUREV.GENET.40.110405.090448
Friedli M., and Trono D., 2015, The developmental control of transposable elements and the evolution of higher species, Annual Review of Cell and Developmental Biology, 31: 429-451.
https://doi.org/10.1146/annurev-cellbio-100814-125514
PMid:26393776
Fukai E., Yoshikawa M., Shah N., Sandal N., Miyao A., Ono S., Hirakawa H., Akyol T., Umehara Y., Nonomura K., Stougaard J., Hirochika H., Hayashi M., Sato S., Andersen S., and Okazaki K., 2022, Widespread and transgenerational retrotransposon activation in inter-and intra-species recombinant inbred populations of Lotus japonicus, The Plant Journal, 111(5): 1397-1410.
https://doi.org/10.1111/tpj.15896
PMid:35792830
Hancks D., and Kazazian H., 2012, Active human retrotransposons: variation and disease, Current Opinion in Genetics and Development, 22(3): 191-203.
https://doi.org/10.1016/j.gde.2012.02.006
PMid:22406018 PMCid:PMC3376660
Hickman A., and Dyda F., 2016, DNA transposition at work, Chemical Reviews, 116(20): 12758-12784.
https://doi.org/10.1021/acs.chemrev.6b00003
PMid:27187082 PMCid:PMC6380494
Howard T., Hayward A., Tordillos A., Fragoso C., Moreno M., Tohme J., Kausch A., Mottinger J., and Dellaporta S., 2014, Identification of the maize gravitropism gene lazy plant1 by a transposon-tagging genome resequencing strategy, PLoS One, 9(1): e87053.
https://doi.org/10.1371/journal.pone.0087053
PMid:24498020 PMCid:PMC3909067
Huang C., Burns K., and Boeke J., 2012, Active transposition in genomes, Annual Review of Genetics, 46: 651-675.
https://doi.org/10.1146/annurev-genet-110711-155616
PMid:23145912 PMCid:PMC3612533
Huda A., and Jordan I., 2009, Epigenetic regulation of mammalian genomes by transposable elements, Annals of the New York Academy of Sciences, 1178(1): 276-284.
https://doi.org/10.1111/j.1749-6632.2009.05007.x
PMid:19845643
Igolkina A., Zinkevich A., Karandasheva K., Popov A., Selifanova M., Nikolaeva D., Tkachev V., Penzar D., Nikitin D., and Buzdin,A., 2019, H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory evolution of open and condensed chromatin landmarks, Cells, 8(9): 1034.
https://doi.org/10.3390/cells8091034
PMid:31491936 PMCid:PMC6770625
Johnson R., and Reznikoff W., 1983, DNA sequences at the ends of transposon Tn5 required for transposition, Nature, 304: 280-282.
https://doi.org/10.1038/304280A0
PMid:6306482
Kawakami K., Largaespada D., and Ivics Z., 2017, Transposons as tools for functional genomics in vertebrate models, Trends in Genetics, 33(11): 784-801.
https://doi.org/10.1016/j.tig.2017.07.006
PMid:28888423 PMCid:PMC5682939
Kruijsbergen I., Hontelez S., Elurbe D., Heeringen S., Huynen M., and Veenstra G., 2017, Heterochromatic histone modifications at transposons in Xenopus tropicalis embryos, Developmental Biology, 426(2): 460-471.
https://doi.org/10.1016/j.ydbio.2016.08.031
PMid:27639284 PMCid:PMC5350053
Li Z., Han L., Luo, Z., and Li L., 2021, Single‐molecule long‐read sequencing reveals extensive genomic and transcriptomic variation between maize and its wild relative teosinte (Zea mays ssp. parviglumis), Molecular Ecology Resources, 22: 272-282
https://doi.org/10.1111/1755-0998.13454
PMid:34157795
Lima-Mendez G., Alvarenga D., Ross K., Hallet B., Melderen L., Varani A., and Chandler M., 2019, Toxin-antitoxin gene pairs found in Tn3 Family transposons appear to be an integral part of the transposition module, mBio, 11(2): 110-128.
https://doi.org/10.1128/mBio.00452-20
PMid:32234815 PMCid:PMC7157771
Lippman Z., May B., Yordan C., Singer T., and Martienssen R., 2003, Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification, PLoS Biology, 1(3): e67.
https://doi.org/10.1371/journal.pbio.0000067
PMid:14691539 PMCid:PMC300680
Mustafin R., and Khusnutdinova E., 2018, The role of transposons in epigenetic regulation of ontogenesis, Russian Journal of Developmental Biology, 49: 61-78.
https://doi.org/10.1134/S1062360418020066
Percharde M., Sultana T., and Ramalho-Santos M., 2020, What doesn't kill you makes you stronger: transposons as dual players in chromatin regulation and genomic variation, BioEssays, 42(4): 1900232.
Perween S., Kumar D., and Kumar A., 2020, A review on transposons and its utilization as genetic tool, International Journal of Current Microbiology and Applied Sciences, 9: 1874-1884.
https://doi.org/10.20546/ijcmas.2020.902.214
Peterson P., 2005, The plant genetics discovery of the century: transposable elements in maize, early beginnings to 1990 [Zea mays L.], Maydica, 50(3): 321-338.
Purugganan M., and Wessler S., 1995, Transposon signatures: species‐specific molecular markers that utilize a class of multiple‐copy nuclear DNA, Molecular Ecology, 4(2): 265-270.
https://doi.org/10.1111/j.1365-294X.1995.tb00218.x
PMid:7735530
Qu S., Desai A., Wing R., and Sundaresan V., 2007, A versatile transposon-based activation tag vector system for functional genomics in cereals and other monocot plants, Plant Physiology, 146(1): 189-199.
https://doi.org/10.1104/pp.107.111427
PMid:17993541 PMCid:PMC2230568
Ramachandran S., and Sundaresan V., 2001, Transposons as tools for functional genomics, Plant Physiology and Biochemistry, 39: 243-252.
https://doi.org/10.1016/S0981-9428(01)01243-8
Ravindran S., 2012, Barbara McClintock and the discovery of jumping genes, Proceedings of the National Academy of Sciences, 109: 20198-20199.
Sandoval-Villegas N., Nurieva W., Amberger M., and Ivics Z., 2021, Contemporary transposon tools: a review and guide through mechanisms and applications of sleeping beauty, piggybac and tol2 for genome engineering, International Journal of Molecular Sciences, 22(10): 5084.
Stitzer M., Anderson S., Springer N., and Ross-Ibarra J., 2019, The genomic ecosystem of transposable elements in maize, PLoS Genetics, 17(10): e1009768.
Su W., Gu X., and Peterson T., 2019, TIR-learner, a new ensemble method for TIR transposable element annotation, provides evidence for abundant new transposable elements in the maize genome, Molecular Plant, 12(3): 447-460.
https://doi.org/10.1016/j.molp.2019.02.008
Tian J., Wang C., Xia J., Wu L., Xu G., Wu W., Li D., Qin W., Han X., Chen Q., Jin W., and Tian F., 2019, Teosinte ligule allele narrows plant architecture and enhances high-density maize yields, Science, 365: 658-664.
https://doi.org/10.1126/science.aax5482
Weil C., and Martienssen R., 2008, Epigenetic interactions between transposons and genes: lessons from plants, Current Opinion in Genetics and Development, 18(2) 188-192.
https://doi.org/10.1016/j.gde.2008.01.015
Xu G.H., Cao J., Wang X., Chen Q., Jin W., Li Z., and Tian F., 2019, Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor, teosinte, Plant Cell, 31: 1990-2009.
Xue C., and Goldenfeld N., 2016, Stochastic predator-prey dynamics of transposons in the human genome, Physical Review Letters, 117(20): 208101.
https://doi.org/10.1103/PhysRevLett.117.208101

. PDF(866KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Zhen Li
Related articles
. Transposons
. Zea genomics
. Genetic architecture
. Genome evolution
. Gene regulation
Tools
. Email to a friend
. Post a comment


